Plot the cosimmr_input data created from cosimmr_load
Source: R/plot.cosimmr_input.R
plot.cosimmr_input.RdThis function creates iso-space (AKA tracer-space or delta-space) plots. They are vital in determining whether the data are suitable for running in a SIMM.
Arguments
- x
An object created via the function
cosimmr_load- tracers
The choice of tracers to plot. If there are more than two tracers, it is recommended to plot every pair of tracers to determine whether the mixtures lie in the mixing polygon defined by the sources
- title
A title for the graph
- xlab
The x-axis label. By default this is assumed to be delta-13C but can be made richer if required. See examples below.
- ylab
The y-axis label. By default this is assumed to be delta-15N in per mil but can be changed as with the x-axis label
- sigmas
The number of standard deviations to plot on the source values. Defaults to 1.
- mix_name
A optional string containing the name of the mixture objects, e.g. Geese.
- colour
If TRUE (default) creates a plot. If not, puts the plot in black and white
- colour_by_cov
if TRUE this allows users to colour the mixtures on the isospace plot by a specified covariate. Defaults to FALSE
- cov_name
The name of the covariate the user wishes to colour the mixture points on the plot by
- ggargs
Extra arguments to be included in the ggplot (e.g. axis limits)
- ...
Not used
Value
isospace plot
Details
It is desirable to have the vast majority of the mixture observations to be inside the convex hull defined by the food sources. When there are more than two tracers (as in one of the examples below) it is recommended to plot all the different pairs of the food sources. See the vignette for further details of richer plots.
See also
See plot.cosimmr_output for plotting the output of a
simmr run. See cosimmr_ffvb for running a cosimmr object once the
iso-space is deemed acceptable.
Examples
# \donttest{
# A simple example with 10 observations, 4 food sources and 2 tracers
data(geese_data_day1)
cosimmr_1 <- with(
geese_data_day1,
cosimmr_load(
formula = mixtures ~ c(1,2,3,2,3,1,2,3,1),
source_names = source_names,
source_means = source_means,
source_sds = source_sds,
correction_means = correction_means,
correction_sds = correction_sds,
concentration_means = concentration_means
)
)
# Plot
plot(cosimmr_1)
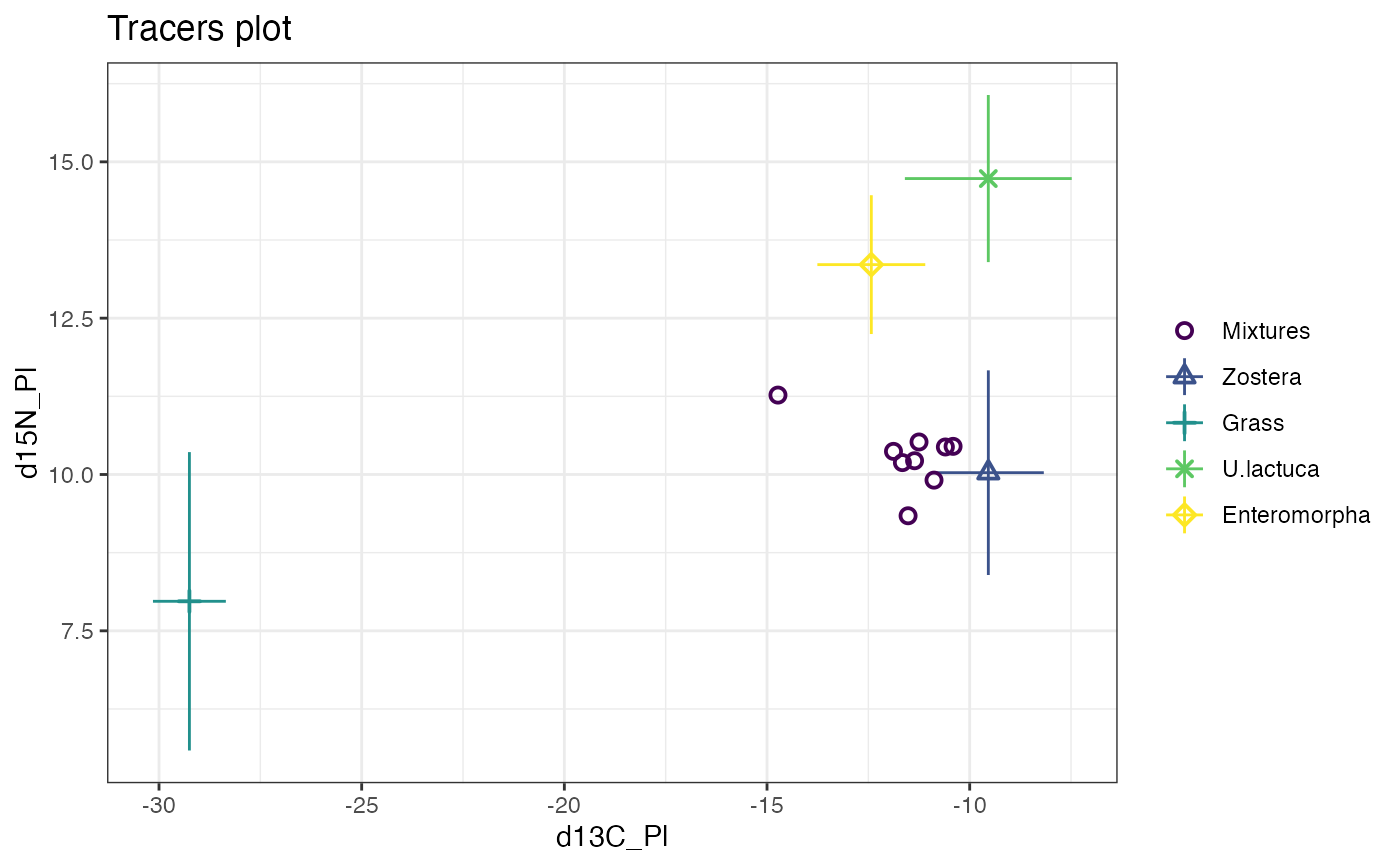 ### A more complicated example with 30 obs, 3 tracers and 4 sources
data(simmr_data_2)
cosimmr_3 <- with(
simmr_data_2,
cosimmr_load(
formula = mixtures ~ 1,
source_names = source_names,
source_means = source_means,
source_sds = source_sds,
correction_means = correction_means,
correction_sds = correction_sds,
concentration_means = concentration_means
)
)
#> Cannot scale when using mixtures ~1
# Plot 3 times - first default d13C vs d15N
plot(cosimmr_3)
### A more complicated example with 30 obs, 3 tracers and 4 sources
data(simmr_data_2)
cosimmr_3 <- with(
simmr_data_2,
cosimmr_load(
formula = mixtures ~ 1,
source_names = source_names,
source_means = source_means,
source_sds = source_sds,
correction_means = correction_means,
correction_sds = correction_sds,
concentration_means = concentration_means
)
)
#> Cannot scale when using mixtures ~1
# Plot 3 times - first default d13C vs d15N
plot(cosimmr_3)
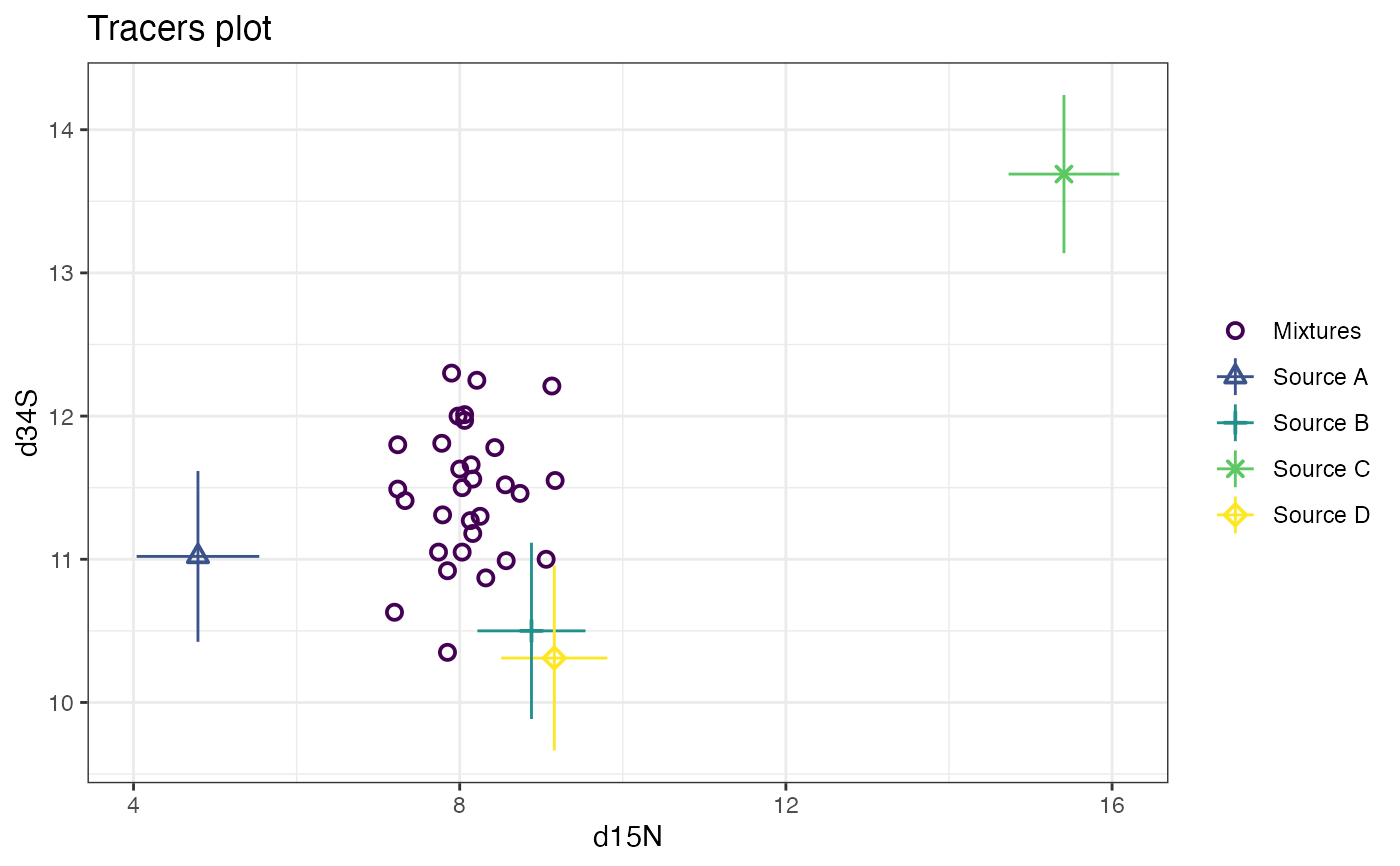
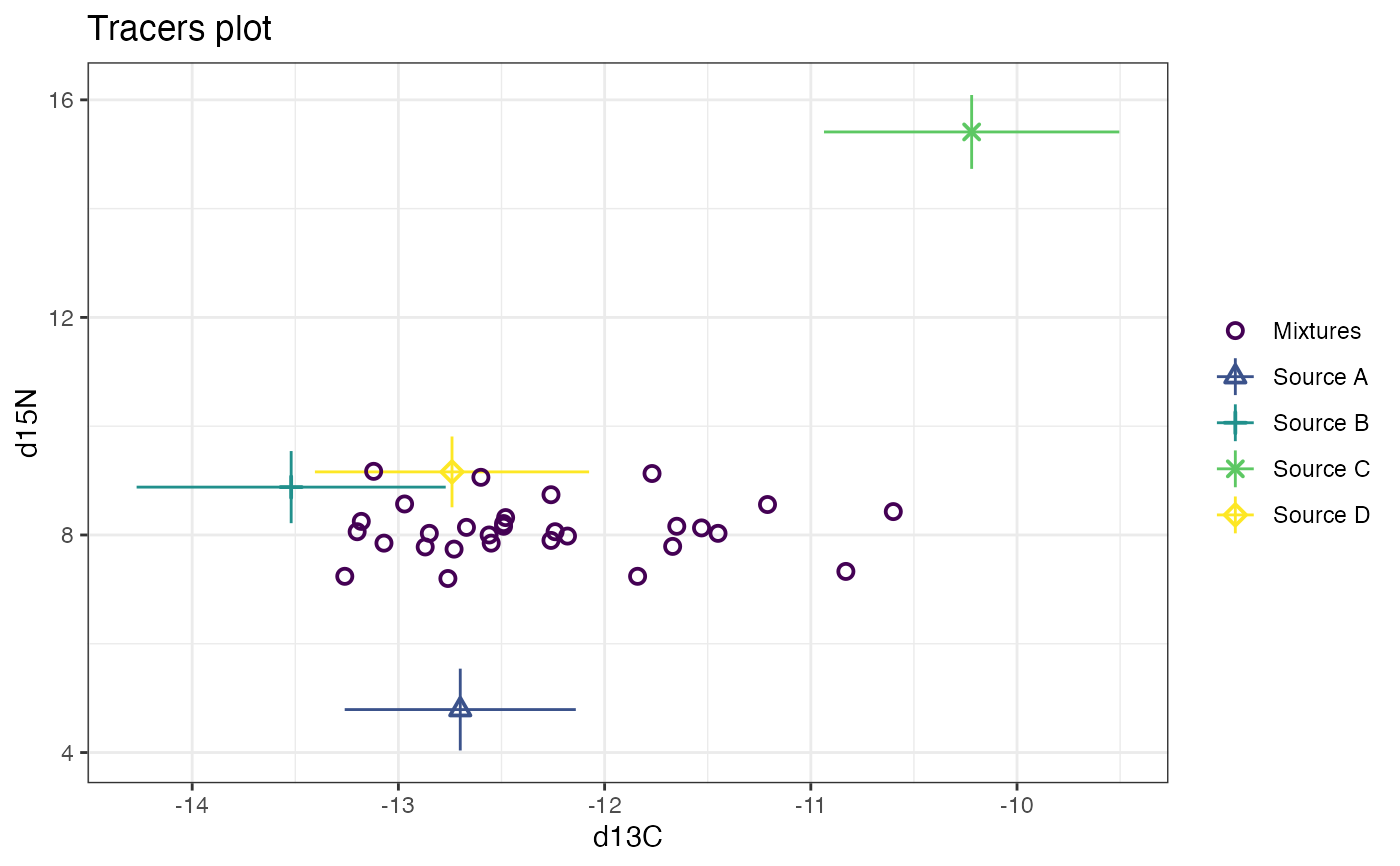 # Now plot d15N vs d34S
plot(cosimmr_3, tracers = c(2, 3))
# Now plot d15N vs d34S
plot(cosimmr_3, tracers = c(2, 3))
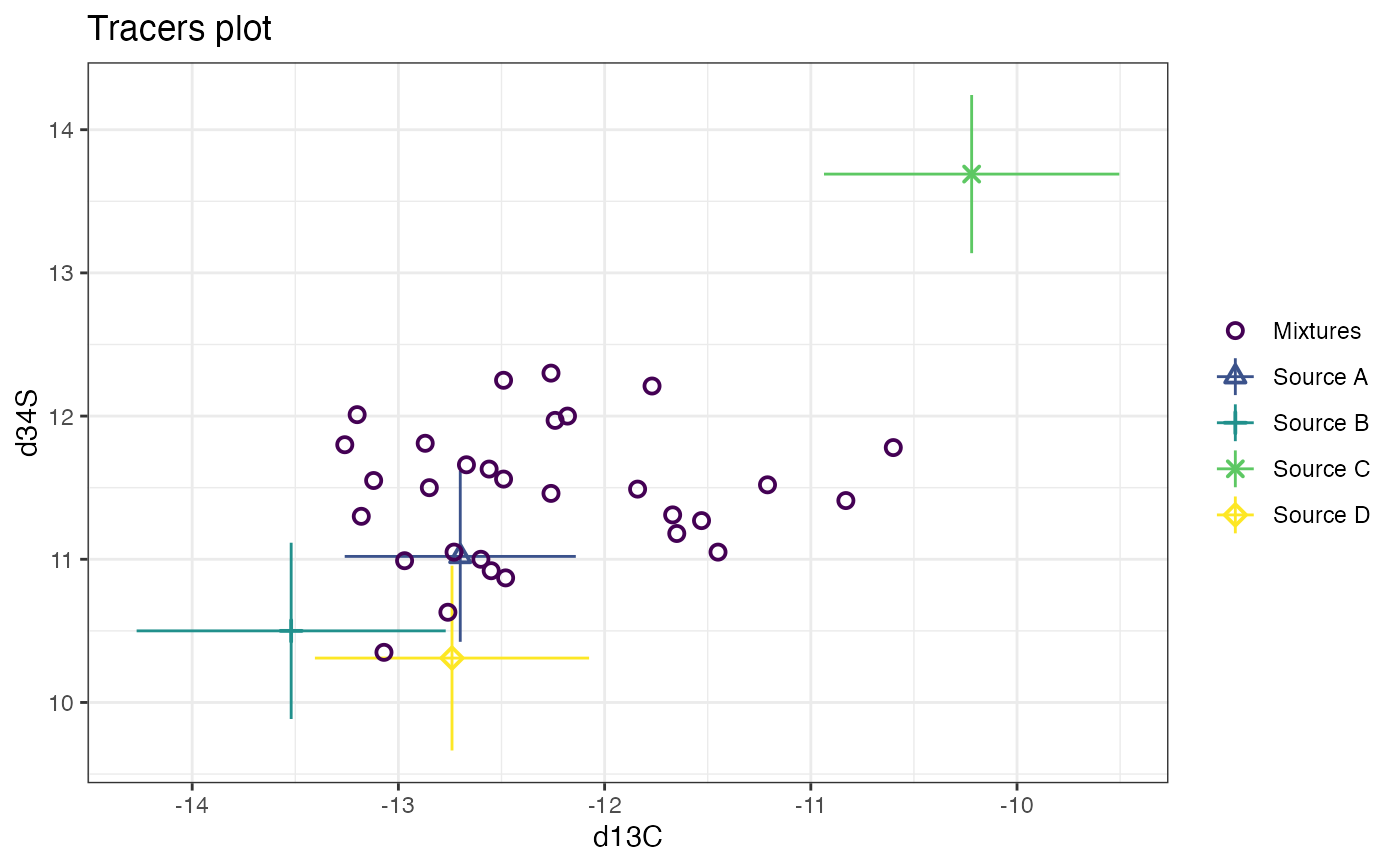
 # and finally d13C vs d34S
plot(cosimmr_3, tracers = c(1, 3))
# and finally d13C vs d34S
plot(cosimmr_3, tracers = c(1, 3))
 # }
# }